Proteome Organization and Cellular Compartmentalization
The Stengel group is studying proteome organization and cellular compartmentalization in health and disease. To this end we are using biochemistry and bioorthogonal chemistry and are developing and applying novel integrated mass spectrometry-based approaches.
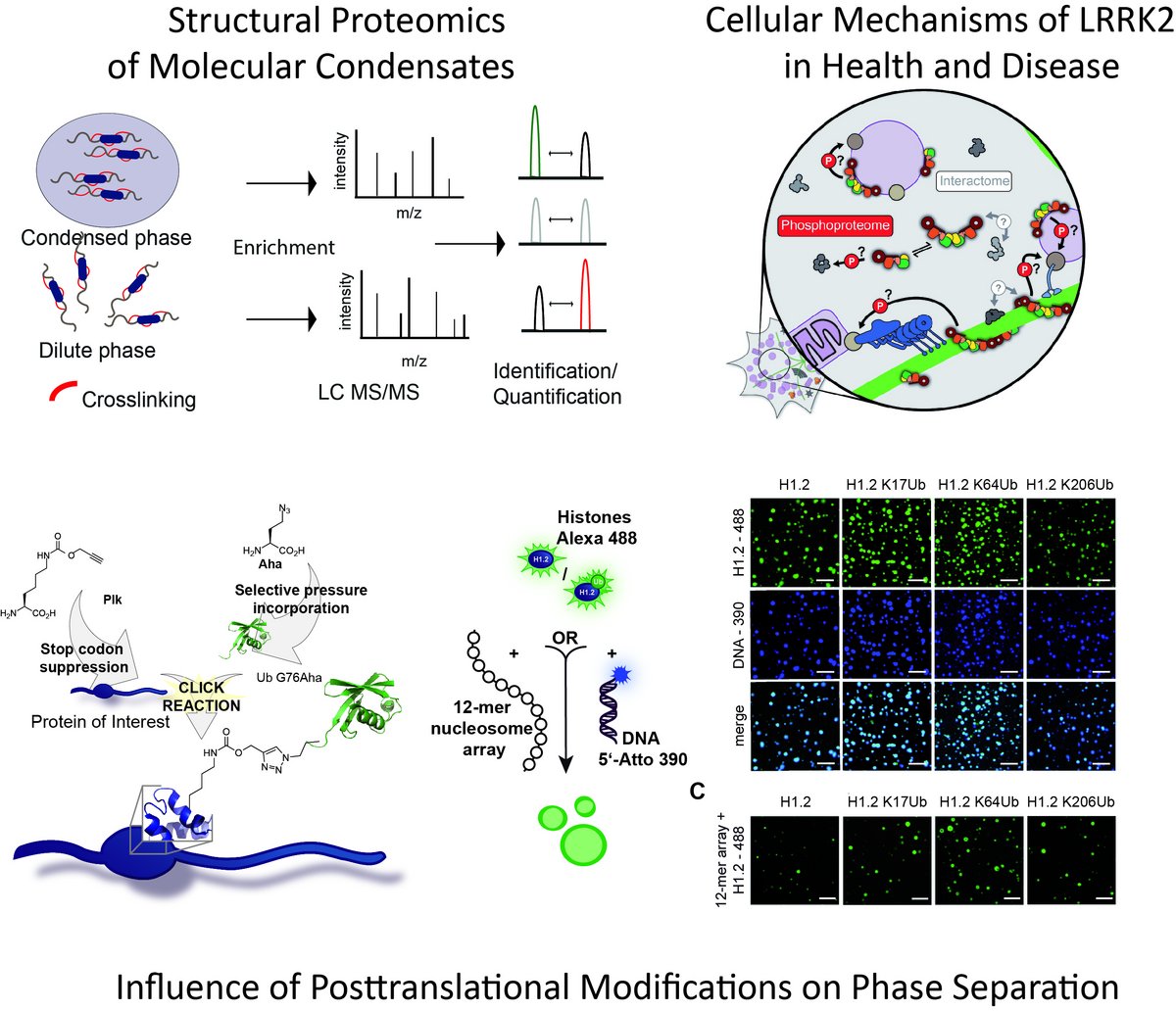
Cells need to control an almost infinite number of processes and biochemical reactions in time and space. A key concept is the organization of the cellular space into functional units and compartments. Cellular compartmentalization scales from single molecules over protein complexes of varying complexity to large membrane surrounded organelles. On the protein and protein complex level posttranslational modifications lead to the generation of functional proteoforms which confer both complexity and dynamics of the system and the emerging concept of liquid-liquid phase separation is increasingly identified as a mechanism to assemble biomolecular condensates that facilitate distinct biochemical reactions and additionally help to organize the functional proteome. Studying proteome organization therefore needs to be able to bridge such different size dimensions. To this end, my group is using biochemistry and bioorthogonal chemistry and is applying and developing integrated mass spectrometry-based approaches in order to address the modular proteome at both the molecular and the systems level and at a timescale that facilitates probing of dynamics.
Specifically, we
1) study how single molecules organize themselves in molecular condensates via phase separation and how this relates to the formation of the modular proteome
(Boczek & Fürsch et al., eLIFE, 2021; Höllmüller & Geigges et al., Nat Commun, 2021)
2) combine bioorthogonal chemistry, biochemistry and MS-based proteomics to study the influence of posttranslational modifications on protein-protein interactions and condensate formation
(Saumer et al., Nucleic Acids Res 2023; Höllmüller & Geigges et al., Nat Commun, 2021; Höllmüller et al, J Proteome Res. 2021; Lutz & Höllmüller et al., Angew. Chem. Int. Ed. 2020; Zhao & Lutz et al., Angew. Chem. Int. Ed. 2017)
3) expand the structural mass spectrometry toolbox
(Chen & Sailer et al., Anal Chem, 2022; Sailer & Jansen et al., Cell Rep., 2022; Boczek & Fürsch et al., eLIFE, 2021; Fürsch et al., Anal Chem, 2021; Huang & Guo & Niedermeier et al, Structure, 2021; Fürsch & Kammer et al., Anal Chem, 2020; Sailer & Offensperger et al., Nat Commun, 2018)
4) investigate cellular mechanisms of LRRK2 in Health and Disease
This project is part of an International Consortium with the goal to understand the function of Leucine Rich Repeat Kinase 2 (LRRK2), the most commonly mutated gene in familial Parkinson’s Disease in normal and diseased cells.
For more information see: (https://parkinsonsroadmap.org/research-network/biology-of-pd-associated-genetics/)
