Research Areas
Precision analytics for Human | Data | Society
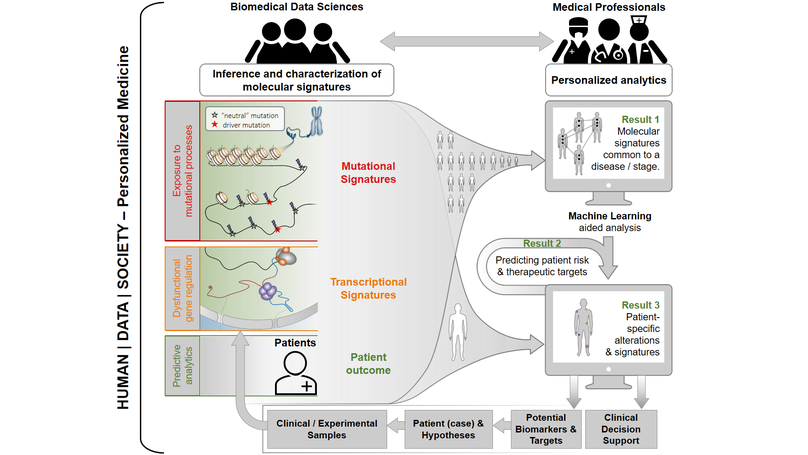
Personalized medicine can improve the lives of patients and clinical professionals lastingly. Thus, developing data science approaches, tools and systems for personalized analytics is one of the aims of our research group, which is a member of the recently established Centre for Human | Data | Society. Currently available therapies are mostly designed for the “average” patient and neglect patient-to-patient heterogeneity. However, these ‘one-size-fits-all’ treatment approaches are not suitable for most complex diseases, such as cancers, as they do not take into account the molecular properties of an individual disease. In our research we develop and apply data science approaches that enable to extract previously unknown molecular signatures and unlock their biomarker potential for personalized medicine. By analyzing the signatures in terms of their occurrence within certain cancer types and their association with disease progression, we develop predictive models that aim to support personalized diagnostics and clinical decision making in the future.
Related Publications
- Gruber AJ, Zavolan M, “Alternative cleavage and polyadenylation in health and disease.” Nature Reviews Genetics. 2019 Oct;20(10):599-614. (doi)
- Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, and van Nimwegen E, “ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs.” Genome Research 2014 May;24(5):869-84. (doi), (Data Science Tool)
- Islam SMA, Díaz-Gay M, Wu Y, Barnes M, Vangara R, Bergstrom EN, He Y, Vella M, Wang J, Teague JW, Clapham P, Moody S, Senkin S, Li YR, Riva L, Zhang T, Gruber AJ, Steele CD, Otlu B, Khandekar A, Abbasi A, Humphreys L, Syulyukina N, Brady SW, Alexandrov BS, Pillay N, Zhang J, Adams DJ, Martincorena I, Wedge DC, Landi MT, Brennan P, Stratton MR, Rozen SG, Alexandrov LB, “Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor.” Cell Genomics. 2022 Nov 9;2(11). (doi), (Data Science Tool)
- Gruber AJ, Schmidt R, Ghosh S, Martin G, Gruber AR, van Nimwegen E, Zavolan M, “Discovery of physiological and cancer-related regulators of 3' UTR processing with KAPAC.” Genome Biology, 2018 Mar 28;19(1):44. (doi), (Data Science Tool)
- Lal A, Galvao Ferrarini M, Gruber AJ, “Investigating the Human Host-ssRNA Virus Interaction Landscape Using the SMEAGOL Toolbox.” Viruses. 2022 Jun 29;14(7):1436. (doi), (Data Science Tool)
- Ferrarini MG, Lal A, Rebollo R, Gruber AJ, Guarracino A, Gonzalez IM, Floyd T, de Oliveira DS, Shanklin J, Beausoleil E, Pusa T, Pickett BE, Aguiar-Pulido V., “Genome-wide bioinformatic analyses predict key host and viral factors in SARS-CoV-2 pathogenesis.” Communications Biology, 2021 May 17;4(1):590. doi: 10.1038/s42003-021-02095-0. (doi)
- Ansari-Pour N, Zheng Y, Yoshimatsu TF, Sanni A, Ajani M, Reynier JB, Tapinos A, Pitt JJ, Dentro S, Woodard A, Rajagopal PS, Fitzgerald D, Gruber AJ, Odetunde A, Popoola A, Falusi AG, Babalola CP, Ogundiran T, Ibrahim N, Barretina J, Van Loo P, Chen M, White KP, Ojengbede O, Obafunwa J, Huo D, Wedge DC, Olopade OI, “Whole-genome analysis of Nigerian patients with breast cancer reveals ethnic-driven somatic evolution and distinct genomic subtypes.” Nature Communucations, 2021 Nov 26;12(1):6946. (doi)
Alternative cleavage and polyadenylation in health and disease
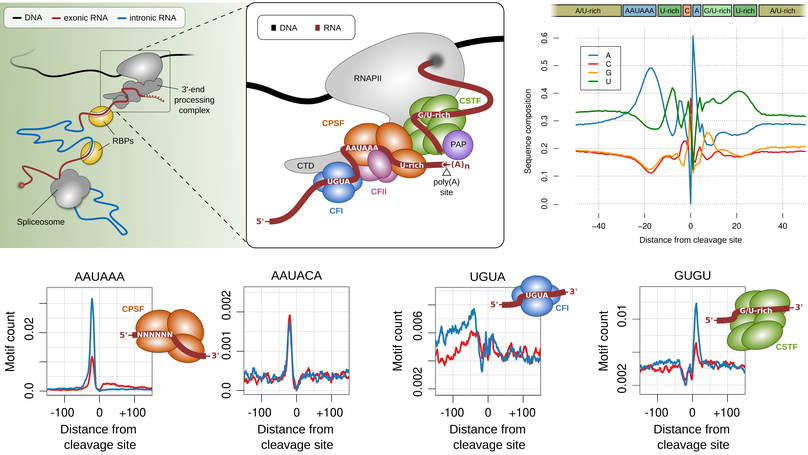
The 3' end of RNA polymerase II transcripts is generated by endonucleolytic cleavage and polyadenylation at 3' end processing sites, also termed poly(A) sites. The processing of 3' end processing sites is mediated by the so called 3' end processing complex, which is a huge machinery that consists out of several subcomplexes (CFI, CPSF, CSTF, CFII) that bind to specific sequence motifs in vicinity to poly(A) sites, the most prominent of which is the so called canonical poly(A) signal (‘AAUAAA’). Most human genes have multiple poly(A) sites and the alternative cleavage and polyadenylation (APA) of these sites gives rise to isoforms that differ in their coding sequence (CDS) and/or their 3' untranslated regions (3' UTRs). The latter harbour cis-regulatory elements that are key regulators of RNA tability, translation and localization.
Identification of molecular cancer subtypes
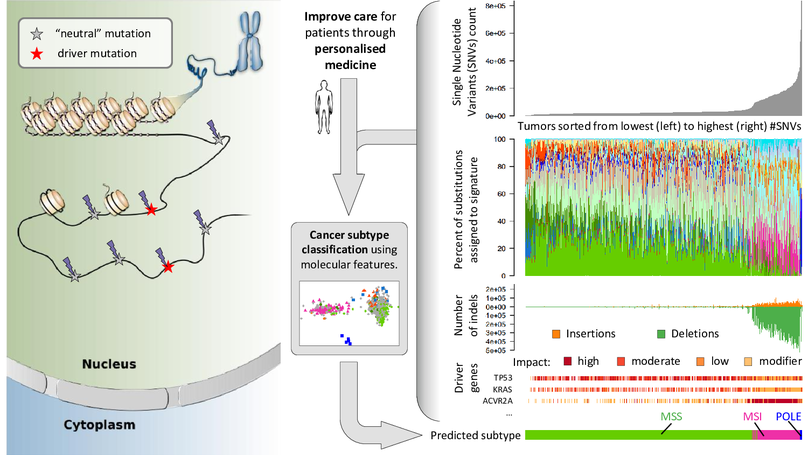
Tumors harbour various molecular features, such as tumor mutation burden (single nucleotide variants and indels), specific driver gene mutations and mutational signatures. We are subtyping cancers based on various molecular alterations at a fine level of detail and investigate their association with clinical outcomes.
