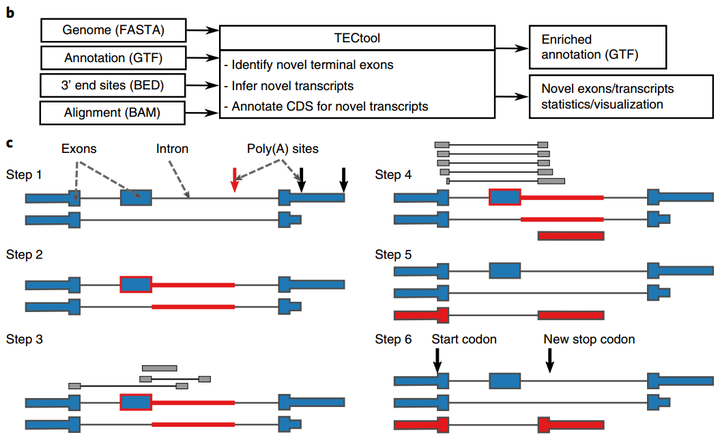
Software, Tools & Databases
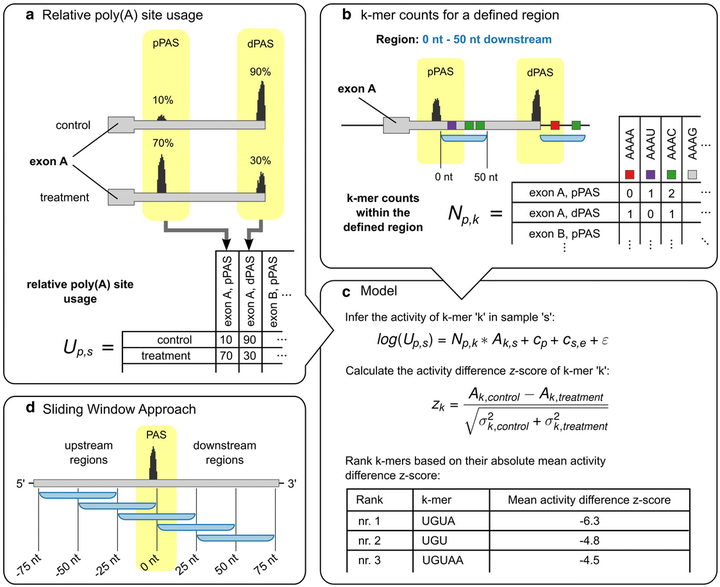
KAPAC
The KAPAC tool enables the identification of sequence motifs that can explain changes in cellular 3' end processing from high-throughput sequencing data.
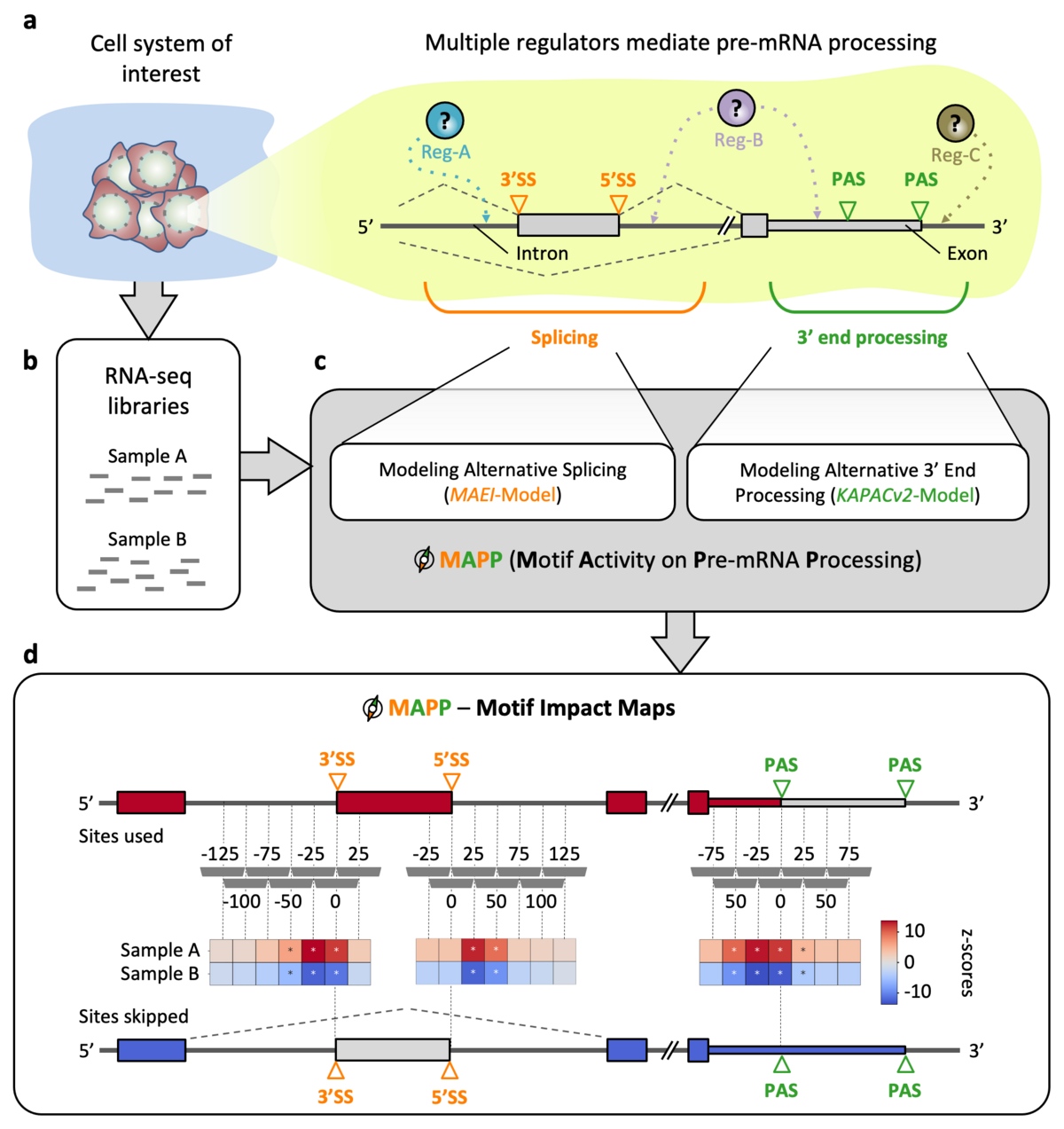
MAPP
To characterize the impact of RNA-binding proteins (RBPs) on splicing and/or polyadenylation (poly(A)) we have developed MAPP (Motif Activity on Pre-mRNA Processing), a fully automated computational tool. MAPP enables the inference of the RBP sequence motif-guided regulation of pre-mRNA processing from standard RNA sequencing (RNA-seq) data.
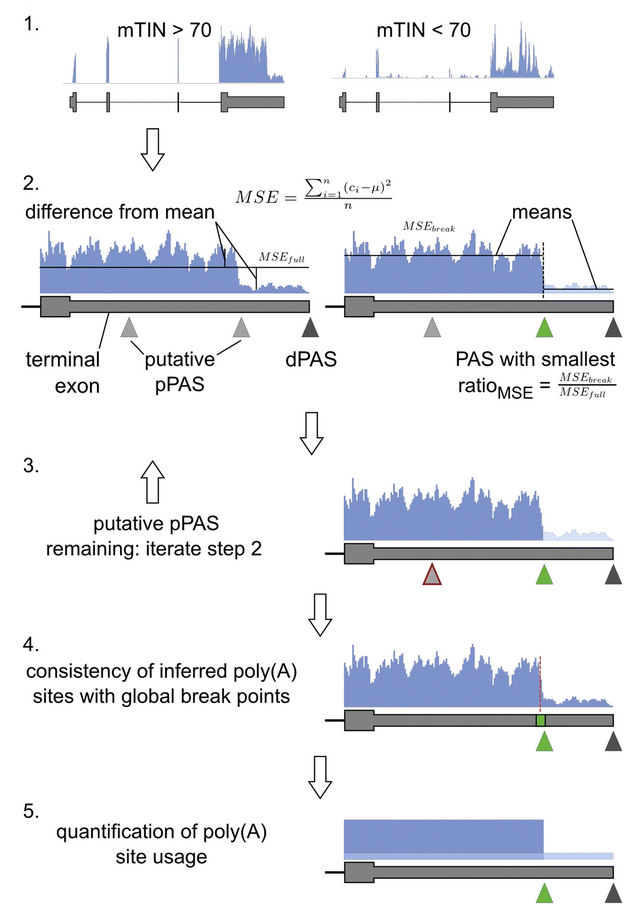
PAQR
PAQR, standing for polyadenylation site usage quantification from RNA sequencing data, allows to quantify 3' end usage from standard RNA sequencing libraries.
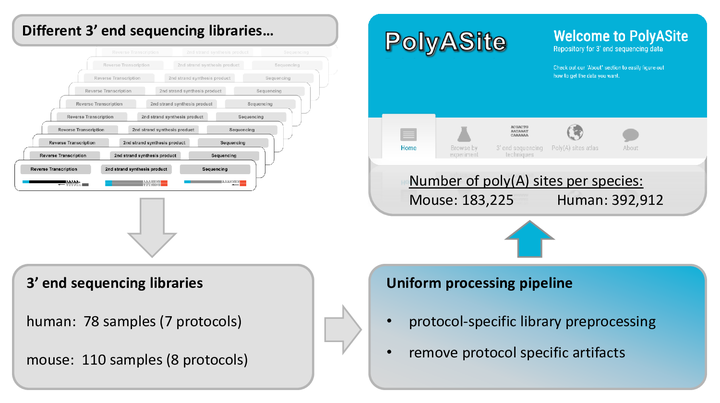
PolyASite
A comprehensive repository for 3' end processing sites based on a uniform analysis of a large number of 3' end sequencing data sets.