SQ and SQDG biodegradation
Sulfoquinovose (SQ, 6-deoxy-6-sulfoglucose) has been known for 50 years as the polar headgroup of the plant sulfolipid SQ-diacylglycerol (SQDG) in the photosynthetic membranes of all higher plants, mosses, ferns, algae, and most photosynthetic bacteria. SQDG is also found in some non-photosynthetic bacteria, and SQ is part of the surface layer of some Archaea. The estimated annual production of SQ is 10,000,000,000 tonnes, thus comprising a major portion of the organo-sulfur in nature, where SQ is degraded by bacteria.
Until today, we revealed three bacterial degradation pathways for SQ, and one biochemical route for anaerobic degradation of SQ to H2S, as detailed in the following.
'Sulfoglycolysis' pathway in Escherichia coli K-12
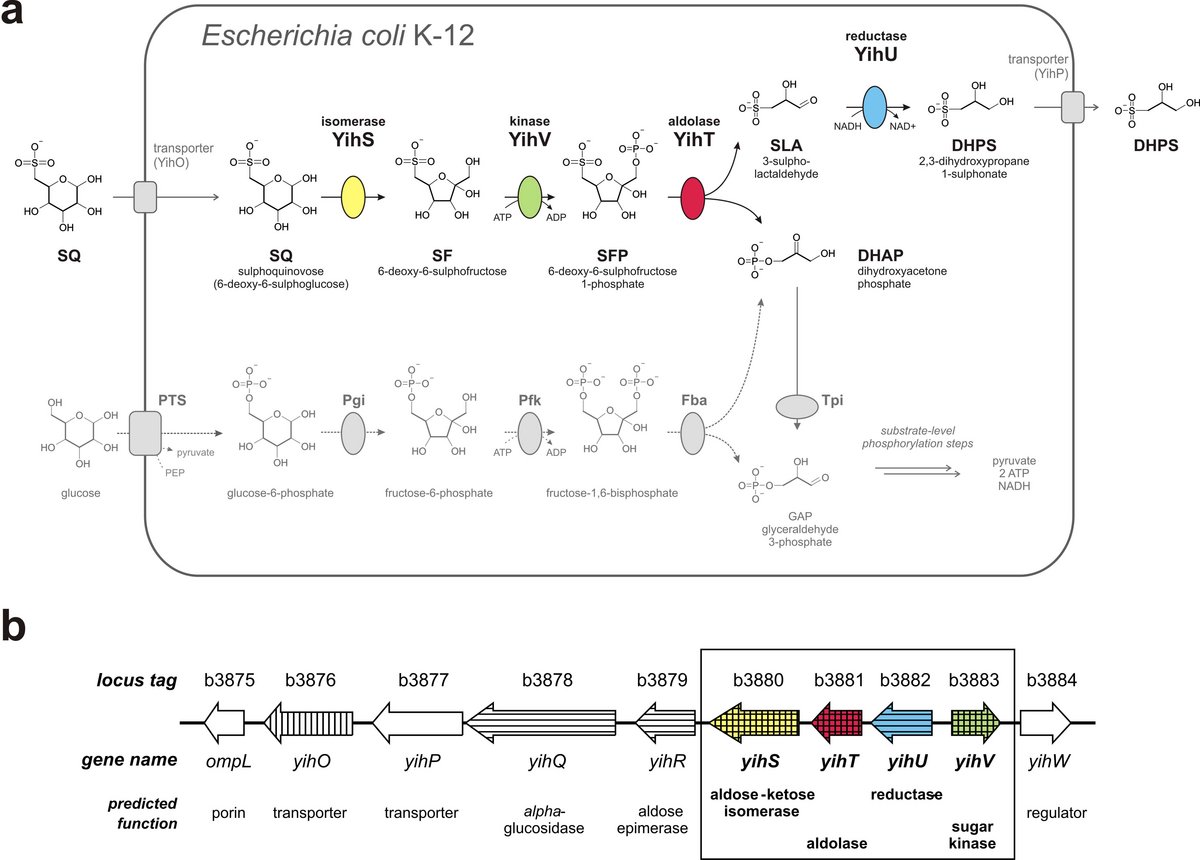
Escherichia coli catabolizes SQ through four newly discovered reactions that we established using purified, heterologously expressed enzymes [see Fig. 1A below]: 6-deoxy-6-sulfoglucose (SQ) isomerase, 6‑deoxy‑6‑sulfofructose (SF) kinase, 6-deoxy-6-sulfofructose-1-phosphate (SFP) aldolase, and 3‑sulfolactaldehyde (SLA) reductase. The pathway yields dihydroxyacetone phosphate (DHAP), which powers growth of E. coli, and the C3-organosulfonate product 2,3-dihydroxypropane-1-sulfonate (DHPS), which is excreted. The enzymes are encoded in a ten-gene cluster [Fig. 1B], which probably encodes also regulation, transport and degradation of the whole sulfolipid.
The sulfoglycolysis gene cluster is present in almost all (>91%) available E. coli genomes, and is widespread in Enterobacteriaceae, such as in genomes of Salmonella, Pantoea and Chronobacter species. Hence, sulfoglycolysis is likely to play a significant role in bacteria in the alimentary tract of all omnivores and herbivores, and in plant pathogens.
'Sulfo-Entner-Doudoroff' pathway in Pseudomonas putida SQ1
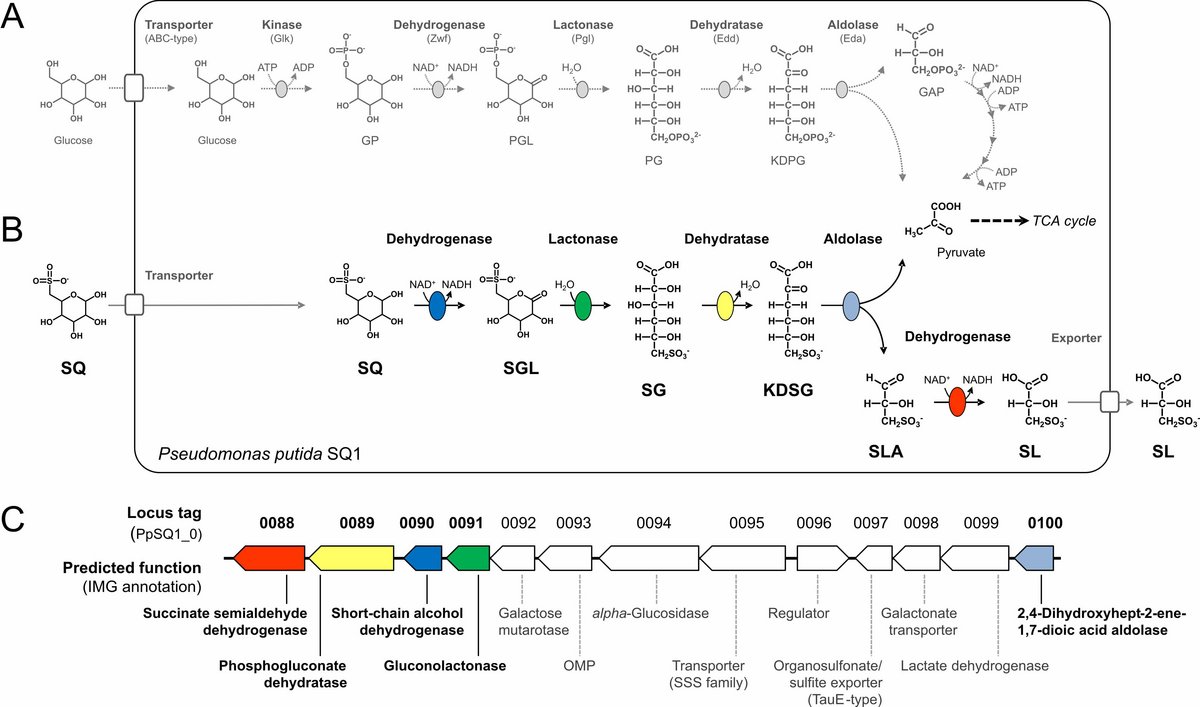
Environmental isolate Pseudomonas putida SQ1 is also able to utilize SQ for growth, and excretes a different C3-organosulfonate, 3‑sulfolactate (SL). We revealed the catabolic pathway for SQ in P. putida SQ1 through differential proteomics and transcriptional analyses, in-vitro reconstitution of the complete pathway by five heterologously produced enzymes, and by identification of all four organosulfonate intermediates by HPLC-MS/MS. The pathway follows a reaction sequence analogous to the Entner-Doudoroff pathway for glucose-6-phosphate [Figures 2A and 2B]: it involves a NAD+-dependent SQ dehydrogenase, 6‑deoxy-6-sulfogluconolactone (SGL) lactonase, 6‑deoxy-6-sulfogluconate (SG) dehydratase, and 2‑keto-3,6-dideoxy-6-sulfogluconate (KDSG) aldolase. The aldolase reaction yields pyruvate, which supports growth of P. putida, and 3‑sulfolactaldehyde (SLA), which is oxidized to SL by an NAD(P)+-dependent SLA dehydrogenase. All five enzymes are encoded in a single gene cluster that includes also genes for, e.g., transport and regulation [Fig. 2C].
Homologous gene clusters were found in genomes of other P. putida strains, in other gamma-proteobacteria, and in beta- and alpha-proteobacteria, for example in genomes of Enterobacteria, Vibrio, Halomonas species, and in typical soil bacteria such as Burkholderia, Herbaspirillum and Rhizobium. Some of these strains are isolated gut symbionts (e.g., the Enterobacteria Hafnia alvei ATCC 51873, Leminorella grimontii DSM5078, and Serratia sp. Ag2) and/or potential pathogens (e.g., Vibrio, Plesiomonas, Halomonas), while other strains represent typical marine or saline (Halomonas, Salinarimonas), and typical freshwater, soil and plant-rhizosphere associated bacteria (e.g., Rhizobium, Ensifer, Herbaspirillum, Microvirga).
6-Deoxy-6-sulfofructose transaldolase pathway for SQ in environmental and intestinal phylum Firmicutes bacteria
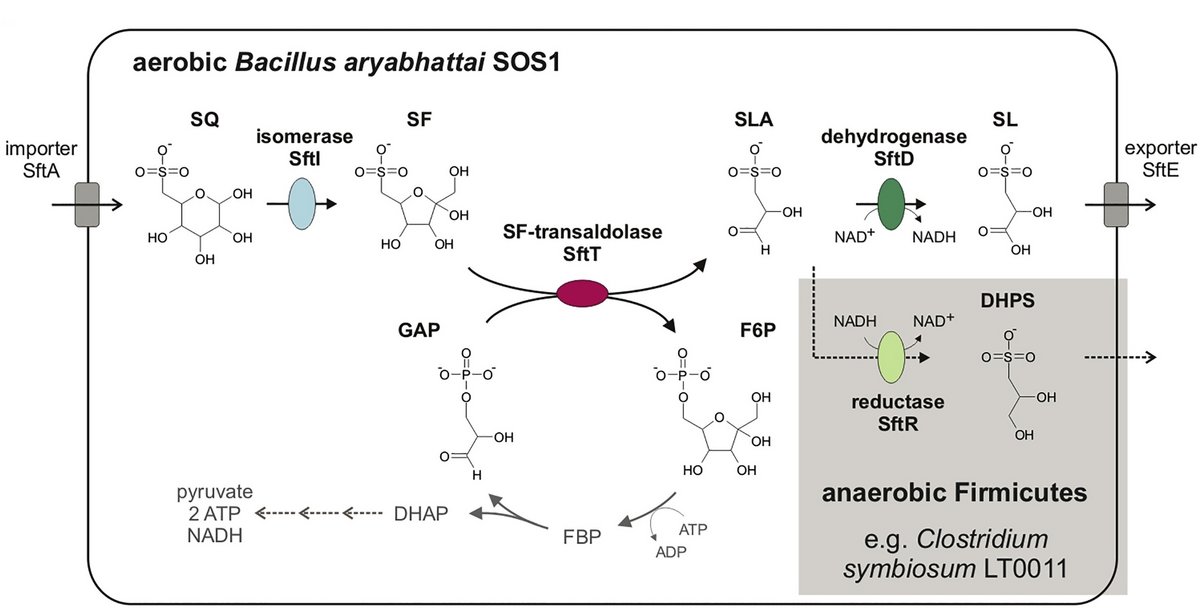
The third pathway for SQ degradation was discovered in a newly isolated, aerobic Bacillus aryabhattai strain, which excretes SL during SQ degradation. It represents the first known SQ-utilizing Gram-positive (phylum Firmicutes) bacterium. Its SQ pathway [Fig. 3] was identified by differential proteomics and reconstituted by heterologously produced enzymes, and all key intermediates were identified by mass spectrometry, using 13C6-isotopically labeled SQ as substrate: It employs an SQ-isomerase, converting SQ to 6-deoxy-6-sulfofructose (SF). A novel transaldolase enzyme cleaves the SF to 3-sulfolactaldehyde (SLA), while the non-sulfonated C3-(glycerone)-moiety is transferred to an acceptor molecule, glyceraldehyde phosphate (GAP), yielding fructose-6-phosphate (F6P).
Additionally, an anaerobic, SQ-fermenting strain, Clostridium symbiosum LT0011, was isolated from human feces, and we demonstrated that this strain and two common human gut bacteria, Enterococcus gilvus DSM15689 and Eubacterium rectale DSM17629, harbor and express this newly discovered pathway during SQ fermentation. In these anaerobic strains, the 3-sulfolactaldehyde (SLA) intermediate is also reduced to DHPS as an additional fermentation step, as with the E. coli pathway [see Fig. 1]. Hence, metabolism of dietary SQ in intestinal microbiomes may be catalyzed by members of the family Enterobacteriaceae via the known sulfogylcolysis pathway [Fig. 1], as well as by members of the phylum Firmicutes via the 6-deoxy-6-sulfofructose transaldolase pathway [Fig. 3]. Further, candidate gene clusters were found in individual genomes of members of other phyla than Firmicutes, that is Fusobacteria, Chloroflexi, Actinobacteria, Spirochaetes, and Thermotogae.
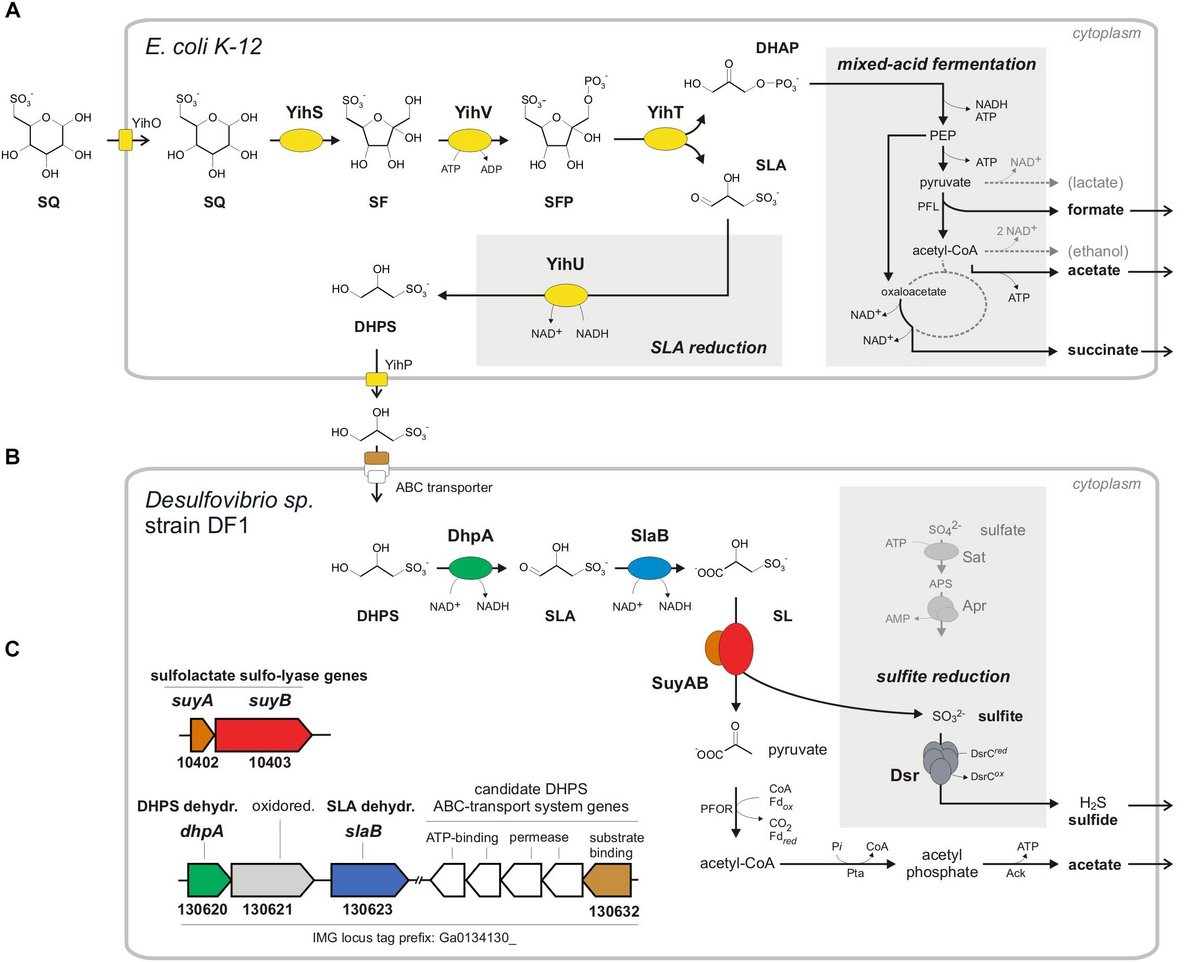
Anaerobic degradation of SQ concomitant with H2S production: Escherichia coli K-12 and Desulfovibrio sp. strain DF1 as co-culture model
SQ is known to be degraded by aerobic bacterial consortia in two tiers via C3-organosulfonates (SL or DHPS) as transient intermediates to CO2, water and sulfate. We demonstrated anaerobic degradation of SQ by bacterial consortia in two tiers to acetate and hydrogen sulfide (H2S), using a laboratory model system constituted of E. coli K-12 and a newly isolated Desulfovibrio sp. strain [Fig. 4]. For the first tier, the E. coli catalyzes a fermentation of SQ to 2,3-dihydroxypropane-1-sulfonate (DHPS), succinate, acetate and formate, thus, a novel type of mixed-acid fermentation. It employs the characterized SQ sulfoglycolysis (SQ Embden-Meyerhof-Parnas) pathway [see Fig. 1], as confirmed by mutational and proteomic analyses. For the second tier, the DHPS-degrading Desulfovibrio sp. isolate catalyzes another novel fermentation, of the DHPS to acetate and H2S. Its DHPS desulfonation pathway was identified by differential proteomics and demonstrated by heterologously produced enzymes: DHPS is oxidized via 3-sulfolactaldehyde (SLA) to 3-sulfolactate (SL) by two NAD+-dependent dehydrogenases (DhpA, SlaB); the SL is cleaved by an SL sulfite-lyase known from aerobic bacteria (SuyAB) to pyruvate and sulfite [Fig. 4B]. The pyruvate is oxidized to acetate, while the sulfite is used as electron acceptor in respiration and reduced to H2S.
In conclusion, anaerobic sulfidogenic SQ degradation was demonstrated as a novel link in the biogeochemical sulfur cycle. Since SQ is also a constituent of the green-vegetable diet of herbivores and omnivores and H2S production in the intestinal microbiome has many recognized and potential contributions to human health and disease, it is important to examine in future the bacterial SQ degradation in the human intestinal microbiome in situ, in relation to H2S production, dietary conditions and human health.