
Junior Group Gamerdinger
Elucidating the molecular interplay of nascent chain processing factors on translating ribosomes
Failure of protein homeostasis poses a substantial burden for cellular health, and the pathogenesis of numerous human diseases, such as Alzheimer´s disease, Parkinson´s disease and Prion disease, are causally linked to protein folding defects. To avoid protein misfolding and aggregation cells invest in a highly complex and interconnected set of mechanisms that affect the levels and conformational stability of proteins. At the core of this protein homeostasis (proteostasis) system are pathways that control the biogenesis, folding, transport and degradation of proteins in the intra- and extracellular space.
Fundamental for maintaining protein homoeostasis in cells is to avoid creating defective proteins during the translation process. The correct folding and localization of newly synthesized proteins is governed by a repertoire of conserved protein biogenesis factors that directly bind to the ribosome and chaperone nascent polypeptide chains as soon as they emerge from the ribosomal tunnel exit (see the figure below). This nascent chain ‘welcoming committee’ regulates multiple co-translational processes including protein modifications, folding, targeting and degradation. Acting at the front of the protein production line, these ribosome-associated factors lead the way in the cellular protein homeostasis network to ensure proteome integrity.
The Figure shows an overview of conserved ribosome-associated protein biogenesis factors. All these factors use the ribosome as a platform to process their substrates in a co-translational manner. How these factors gain regulated access to nascent chains and how they affect each others binding specificity is so far not well understood.
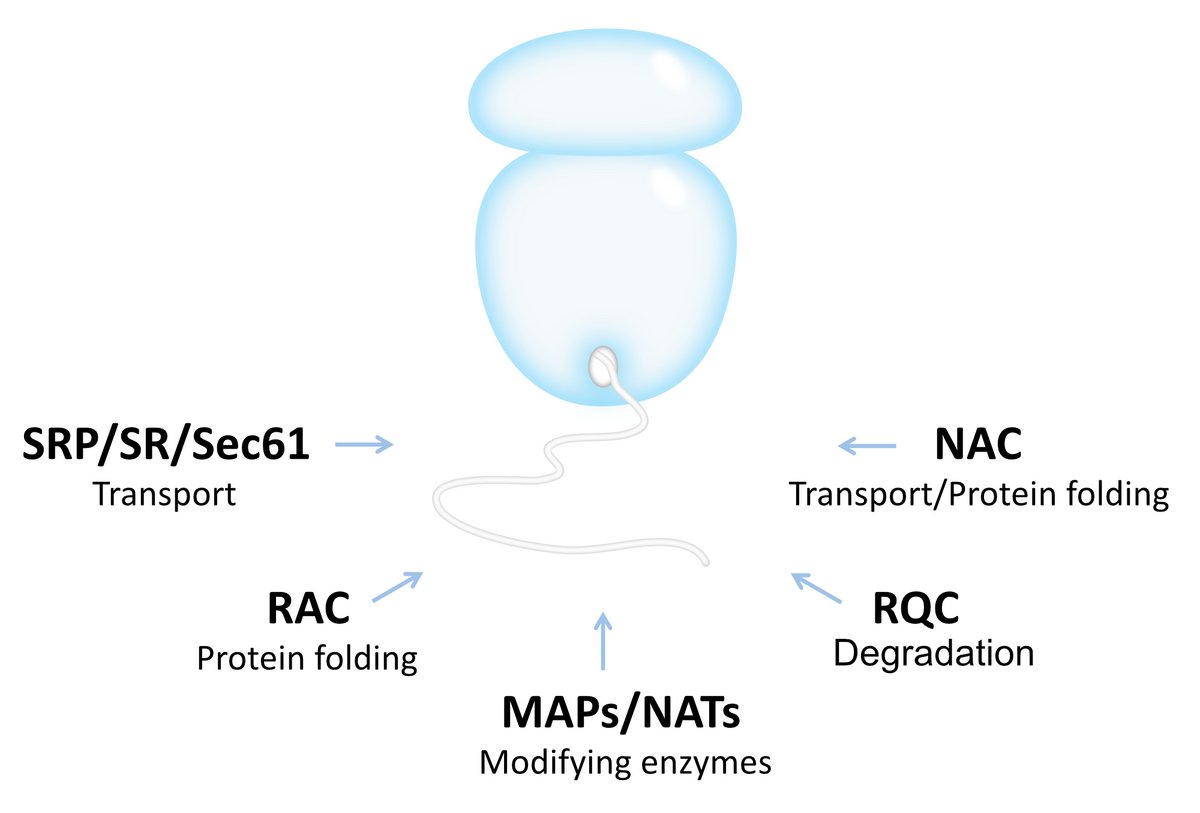
The timely and accurate selection of nascent polypeptides into the correct biogenesis pathway is an essential key process to avoid protein misfolding and aggregation. By using the ribosome as a platform, the different nascent chain-processing systems are enabled to assist protein biogenesis processes at the earliest possible time. However, many of these factors use overlapping binding sites on ribosomes and an emerging question is how these factors gain regulated access to nascent polypeptides.
Our research group is interested in investigating the function and molecular interplay of different protein biogenesis factors that bind to the ribosome exit site. We could recently show that one of these factors, the nascent polypeptide-associated complex (NAC), is crucial to regulate the ribosomal access of the Sec61 translocon complex (Gamerdinger et al. Science, 2015). Based on this finding, we follow the hypothesis that NAC, the only exit site-binding factor that is expressed in equimolar concentration relative to ribosomes, is the central regulator that orchestrates the access and thus the specificity of substrate selection of nascent chain processing factors, thereby taking a leading role in guarding the early life of a protein.
To elucidate the function of ribosome-associated protein biogenesis factors we use state-of-the-art biochemical and molecular genetic approaches. As an in vivo animal model we primarily employ the model organism Caenorhabditis elegans that allows not only the investigation of general but also tissue-specific and potential age- and disease-associated alterations of co-translational processing events at the ribosome exit site.
Selected Publications
Gamerdinger, M., Kobayashi, K., Wallisch, A., Kreft, S.G., Sailer, C., Schlömer, R., Sachs, N., Jomaa, A., Stengel, F., Ban, N., and Deuerling, E. (2019) Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol. Cell 75, 996-1006, short recommended in F1000Prime as being of special significance in its field
Shen, K., Gamerdinger, M., Chan, R., Gense, K., Martin, E.M., Sachs, N., Knight, P.D., Schlömer, R., Calabrese, A.N., Stewart, K.L., Leiendecker, L., Baghel, A., Radford, S.E., Frydman, J., and Deuerling, E. (2019) Dual role of ribosome-binding domain of NAC as a potent suppressor of protein aggregation and aging-related proteinopathies. Mol. Cell, epub ahead of print, doi: 10.1016/j.molcel.2019.03.012. short | full
Wurmthaler, L.A., Sack, M., Gense, K., Hartig, J., and Gamerdinger, M. (2019) A tetracycline-dependent ribozyme switch allows conditional induction of gene expression in Caenorhabditis elegans. Nat. Commun. 10, 491 short | full
Gamerdinger, M. (2016) Protein quality control at the ribosome: focus on RAC, NAC and RQC. Essays Biochem. 60, 203-212 short | full
Gamerdinger, M., Hanebuth, M.A., Frickey, T., and Deuerling, E. (2015) The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 348, 201-207 short | full
Gamerdinger, M., and Deuerling, E. (2014) Trigger Factor flexibility. Science 344:590-591 full
Gamerdinger, M., and Deuerling, E. (2012) Macrolides: the plug is out. Cell 151, 469. short | full
